
Ayuda de traducción - Aide de traduction - Assistenza di traduzione - Übersetzungsunterstützung
Last modified:
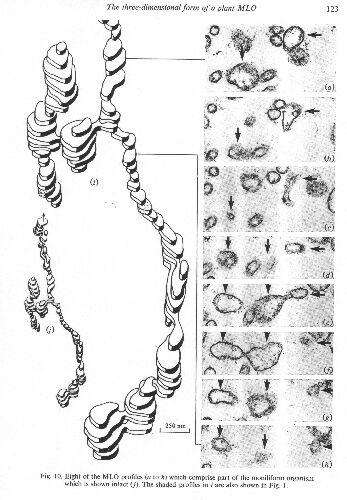
Pathogen When the disease was first named Lethal Yellowing (Nutman & Roberts, 1955) it was thought to be a virus. In 1972 three independent laboratories, each using material sampled in Jamaica, simultaneously indentified MLO by transmission EM (Beakbane et al, 1972; Heinze et al, 1972; Plavsic-Banjac et al, 1972). Subsequently, Waters & Hunt (1980) revealed the three-dimensional form of the coconut MLO.
Now, MLO is reclassified as phytoplasma (Lee at al, 1998). The following information was contributed by Nigel Harrrison http://groups.yahoo.com/group/CICLY/message/305
Sequencing and phylogenetic analysis of 16S rRNA genes cloned by conventional means or after amplification by PCR have clearly demonstrated that phytoplasmas comprise a unique monophyletic clade of unculturable, pleomorphic organisms within the class Mollicutesthat are most closely related to the genus Acholeplasma rather than the Mycoplasma. Based on these findings, the term mycoplasma-like organism (MLO) has been abandoned in favor of the generic name of Phytoplasma, a designation that clearly reflects their plant host association. This name has since been widely adopted to refer to these plant pathogens. In addition, phylogenetic analysis of 16S rRNA sequences was subsequently proposed and adopted as a framework on which to build a formal taxonomic classification of the phytoplasmas. Within the phytoplasma clade, as many as 20 subclades (primary groups) have been resolved by these analyses. Similar groups and numerous subgroupings of phytoplasmas have also been delineated by restriction fragment length polymorphism (RFLP) analysis of amplified 16S rDNA and supported by sequence variation in additional, less-conserved evolutionary markers that include ribosomal protein (rp) genes, the 16-23S rRNA intergenic region, and the gene encoding elongation factor Tu. The taxonomic rank of species has been proposed to describe each of the primary phylogenetically coherent phytoplasma groups. The International Committee on Systematic Bacteriology has implemented a scheme for assigning incompletely described prokaryotes such as phytoplasmas to the provisional status Candidatus. Under these guidelines, species names have been formally assigned to five phytoplasma taxa thus far. These are: Phytoplasma aurantifolia,P. australiense, P. australasia, P. fraxini, and P. japonicum. Further speciation is sure to follow.
One of the primary groups defined by molecular classifications is the palm lethal (LY) yellowing group which was designated as group 16SrIV according to the RFLP-based classification scheme of Lee et al. (1998), and as subclade (vii) based on phylogenetic analysis of 16S rDNA sequences (Gundersen et al., 1994). Represented solely by a Florida strain of the LY phytoplasma in these earlier classifications, phytoplasmas associated with decline of coconut (strain LDY, Genbank Accesion No. U18753) and yellows disease of the cyclanth, ‘palma jipi’ (Carludovica palmata Ruiz & Pavon), (strain CPY, Accession No. AF237615), have since been identified and added to this group (Tymon et al, 1998; Cordova, et al., 2000).
Through ongoing collaborative research between our group at Ft. Lauderdale and Carlos Oropeza’s group at the Centro de Investigacion Cientifica de Yucatan (CICY) in Merida, we have recently identified other phytoplasma strains in association with cocount palm displaying leaf yellowing syndromes in southern Mexico that are distinct from the LY phytoplasma. Characterization employing PCR amplification, cloning, RFLP and sequencing of their respective 16S rRNA genes has revealed four previoulsy undescribed, albeit interrelated phytoplasma strains. A synopsis of our most recent findings from this research includes: (a) all four newly recognized coconut-associated strains have been detected in palms in LY-free areas of Mexico; (b) infected tall-type coconut palms exhibit prominent leaf yellowing symptoms easily confused with LY; (c) all four strains are also members of the LY phytoplasma group according to phylogenetic analysis of 16S rDNA; (d) are most similar to strains LDY and CPY rather than to strains associated with typical LY; (e) and, as was previously found for strains LDY and CPY, none of these newest strains are detectable by PCR employing LY phytoplasma-specific, non-ribosomal primers LYF1/R1.
Several other salient points pertinent to the molecular characterization of these strains by RFLP analysis of PCR amplified rDNA are as follows: Characteristically, these strains each contain a single BamHI restriction site in their respective 16S rRNA gene sequence. So far, the only other strains known to possess this feature are phytoplasmas infecting coconut palms in western and eastern Africa. Secondly, LY group strains can be differentiated one from another by AluI digestion of their respective amplified 16S rDNA products. Thirdly, phytoplasmas are known to contain two rRNA operons (Schneider & Seemuller, 1994) and sequence heterogeneity between rRNA operons is evident in most, but not all, LY group strains.
Sequence disparities between the two copies of the 16S rRNA gene in a coconut-infecting strain of the LY phytoplasma were minor and amounted to a total of two single base substitutions only. However, one of these substitutions abolishes a HinfI restriction site in of these genes. Consequently, it is possible to clone and distinguish both gene copies by RFLP analysis using this enzyme. Collectively, RFLP rDNA profiles of Florida LY strains generated by separate digestion with fourteen restriction enzymes match those of LY phytoplasma strains from Mexico, Belize and Honduras. However, all Jamaica LY strains analyzed by this method were unique as HinfI RFLP profiles indicated a lack of rRNA interoperon sequence heterogeneity. Thus, Jamaican strains possess either two identical 16S rRNA genes or a single rRNA operon only. A subtle yet consistent difference. Whether this disparity is an indicator of biologically important differences between LY phytoplasma strains within and between different geographic localities is presently unknown.
In terms of the overall picture concerning LY disease several implications may be drawn from this work. With the risk of overstating the obvious, coconut is a congenial phytoplasma host. Secondly, there are phytoplasmas infecting coconut palms in the Neotropics other than the LY agent. The cumulative evidence indicates that distribution of some, if not all, of the latter strains seemingly predate the arrival of LY, at least in Mexico. These additional strains are associated with leaf yellowing syndromes (LYS) not unlike LY but, most noticeably, affected palms lack the prominent fruit and flower symptoms typical of LY. Field observations also suggest epidemiological differences between coconut LYS and LY disease. It seems that outbreaks of LYS are limited, appearing as scattered foci involving just a few palms with little propensity for further secondary spread to neighboring palms. As such, it is possible that previous occurrence of LYS may have been written off to other causes such as red-ring or perhaps nutritional disorders. The fact that these syndromes did not escape attention in this instance is likely due to a heightened regional awareness and concern surrounding LY due to its continued spread into previously unaffected areas of southern Mexico. Differences in patterns of spread suggest the involvement of vector insects other than the LY vector planthopper Myndus crudus in transmission of LYS agents.
If other vector species are involved, then the next question is whether or not these additional species are also capable of transmitting LY. Lack of abundance of M. crudus on palms in the coastal Yucatan peninsula relative to the abundance and diversity of other potential vector species has long fueled speculation about the existence of other LY vectors in this region. If true, this is not good news in terms of future spread of LY. From a scientific standpoint, the recalcitrant nature of M. crudus has made this species all but impractical for use in vector transmission studies for practical evaluation coconut germplasm for LY resistance. Ultimately, such biological data is the key to resolving issues about differences in virulence amongst phytoplasma strains and an efficient predictor of long term cultivar or hybrid performance. In light of recent developments, vector identification remains a priority area for further research. Certainly it would be very beneficial to have a species conducive to experimental manipulation and transmission studies.
Literature Cited
Cordova, I., Oropeza, C., Almeyda, H. & N. A. Harrison. 2000. First report of a phytoplasma-associated leaf yellowing syndrome of "Palma Jipi" (Carloduvica palmata) plants in southern México. Plant Disease 84: 807.
Gundersen, D. E., Lee, I.-M., Rehner, S. A., Davis, R. E. & D. T. Kingsbury. 1994. Phylogeny of mycoplasma-like organisms (phytoplasmas): a basis for their classification. Journal of Bacteriology 176: 5244-5254.
Lee, I-M., Gundersen-Rindal, D. E., Davis, R. E. & I. M. Bartoszyk. 1998. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rDNA and ribosomal protein gene sequences. International journal of Systematic Bacteriology 48: 1153-1169.
Schneider, B. & E. Seemuller. 1994. Presence of two sets of ribosomal genes in phytopathogenic mollicutes. Applied and Environmental Microbiology 60: 3409-3412.
Tymon, A. M., Jones, P. & N. A. Harrison. 1998. Phylogenetic relationships of coconut phytoplasmas and the development of specific oligonucleotide PCR primers. Annals of Applied Biology 132: 437-452.
This page is under continuous review. If you have an idea or an opinion to improve the contents of the page or the site, tell the editor. If you disagree with anything, say so. If you don't see or get a satisfactory response in a reasonable time contact other participants. At all time keep in contact with other individuals; this site is not a substitute for person to person contact.